MCP for the open-genes
opengenes-mcp
MCP (Model Context Protocol) server for OpenGenes database
This server implements the Model Context Protocol (MCP) for OpenGenes, providing a standardized interface for accessing aging and longevity research data. MCP enables AI assistants and agents to query comprehensive biomedical datasets through structured interfaces.
The server automatically downloads the latest OpenGenes database and documentation from Hugging Face Hub (specifically from the opengenes folder), ensuring you always have access to the most up-to-date data without manual file management.
The OpenGenes database contains:
- lifespan_change: Experimental data about genetic interventions and their effects on lifespan across model organisms
- gene_criteria: Criteria classifications for aging-related genes (12 different categories)
- gene_hallmarks: Hallmarks of aging associated with specific genes
- longevity_associations: Genetic variants associated with longevity from population studies
If you want to understand more about what the Model Context Protocol is and how to use it more efficiently, you can take the DeepLearning AI Course or search for MCP videos on YouTube.
Usage Example
Here's how the OpenGenes MCP server works in practice with AI assistants:
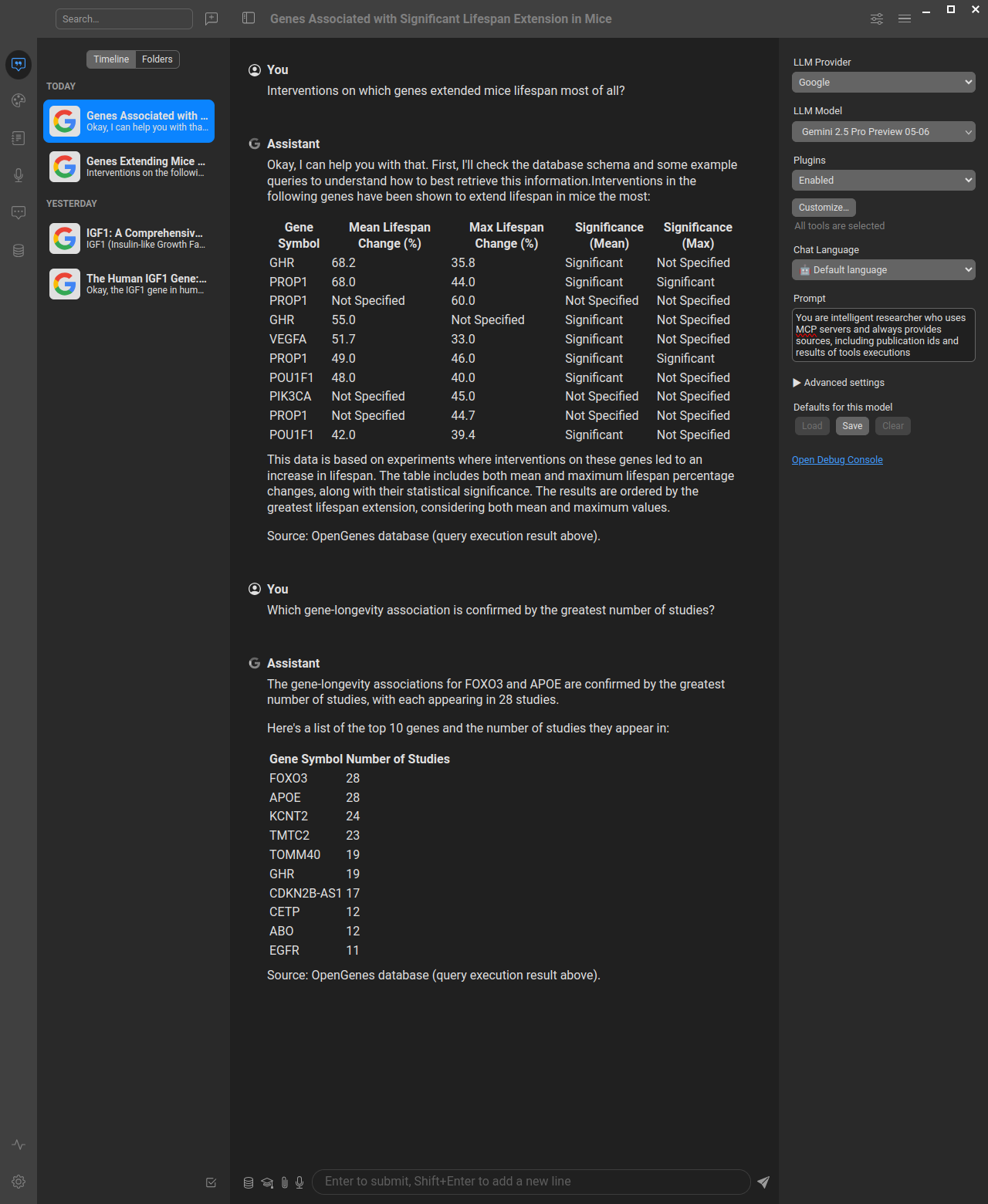
Example showing how to query the OpenGenes database through an AI assistant using natural language, which gets translated to SQL queries via the MCP server. You can use this database both in chat interfaces for research questions and in AI-based development tools (like Cursor, Windsurf, VS Code with Copilot) to significantly improve your bioinformatics productivity by having direct access to aging and longevity research data while coding.
About MCP (Model Context Protocol)
MCP is a protocol that bridges the gap between AI systems and specialized domain knowledge. It enables:
- Structured Access: Direct connection to authoritative aging and longevity research data
- Natural Language Queries: Simplified interaction with specialized databases through SQL
- Type Safety: Strong typing and validation through FastMCP
- AI Integration: Seamless integration with AI assistants and agents
Data Source and Updates
The OpenGenes MCP server automatically downloads data from the longevity-genie/bio-mcp-data repository on Hugging Face Hub. This ensures:
- Always Up-to-Date: Automatic access to the latest OpenGenes database without manual updates
- Reliable Distribution: Centralized data hosting with version control and change tracking
- Efficient Caching: Downloaded files are cached locally to minimize network requests
- Fallback Support: Local fallback files are supported for development and offline use
The data files are stored in the opengenes subfolder of the Hugging Face repository and include:
open_genes.sqlite- The complete OpenGenes databaseprompt.txt- Database schema documentation and usage guidelines
Available Tools
This server provides three main tools for interacting with the OpenGenes database:
opengenes_db_query(sql: str)- Execute read-only SQL queries against the OpenGenes databaseopengenes_get_schema_info()- Get detailed schema information including tables, columns, and enumerationsopengenes_example_queries()- Get a list of example SQL queries with descriptions
Available Resources
resource://db-prompt- Complete database schema documentation and usage guidelinesresource://schema-summary- Formatted summary of tables and their purposes
Quick Start
Installing uv
# Download and install uv
curl -LsSf https://astral.sh/uv/install.sh | sh
# Verify installation
uv --version
uvx --version
uvx is a very nice tool that can run a python package installing it if needed.
Running with uvx
You can run the opengenes-mcp server directly using uvx without cloning the repository:
# Run the server in streamed http mode (default)
uvx opengenes-mcp
Other uvx modes (STDIO, HTTP, SSE)
STDIO Mode (for MCP clients that require stdio, can be useful when you want to save files)
# Or explicitly specify stdio mode
uvx opengenes-mcp stdio
HTTP Mode (Web Server)
# Run the server in streamable HTTP mode on default (3001) port
uvx opengenes-mcp server
# Run on a specific port
uvx opengenes-mcp server --port 8000
SSE Mode (Server-Sent Events)
# Run the server in SSE mode
uvx opengenes-mcp sse
In cases when there are problems with uvx often they can be caused by clenaing uv cache:
uv cache clean
The HTTP mode will start a web server that you can access at http://localhost:3001/mcp (with documentation at http://localhost:3001/docs). The STDIO mode is designed for MCP clients that communicate via standard input/output, while SSE mode uses Server-Sent Events for real-time communication.
Note: Currently, we do not have a Swagger/OpenAPI interface, so accessing the server directly in your browser will not show much useful information. To explore the available tools and capabilities, you should either use the MCP Inspector (see below) or connect through an MCP client to see the available tools.
Configuring your AI Client (Anthropic Claude Desktop, Cursor, Windsurf, etc.)
We provide preconfigured JSON files for different use cases:
- For STDIO mode (recommended): Use
mcp-config-stdio.json - For HTTP mode: Use
mcp-config.json - For local development: Use
mcp-config-stdio-debug.json
Configuration Video Tutorial
For a visual guide on how to configure MCP servers with AI clients, check out our configuration tutorial video for our sister MCP server (biothings-mcp). The configuration principles are exactly the same for the OpenGenes MCP server - just use the appropriate JSON configuration files provided above.
Inspecting OpenGenes MCP server
Using MCP Inspector to explore server capabilities
If you want to inspect the methods provided by the MCP server, use npx (you may need to install nodejs and npm):
For STDIO mode with uvx:
npx @modelcontextprotocol/inspector --config mcp-config-stdio.json --server opengenes-mcp
For HTTP mode (ensure server is running first):
npx @modelcontextprotocol/inspector --config mcp-config.json --server opengenes-mcp
For local development:
npx @modelcontextprotocol/inspector --config mcp-config-stdio-debug.json --server opengenes-mcp
You can also run the inspector manually and configure it through the interface:
npx @modelcontextprotocol/inspector
After that you can explore the tools and resources with MCP Inspector at http://127.0.0.1:6274 (note, if you run inspector several times it can change port)
Integration with AI Systems
Simply point your AI client (like Cursor, Windsurf, ClaudeDesktop, VS Code with Copilot, or others) to use the appropriate configuration file from the repository.
Repository setup
# Clone the repository
git clone https://github.com/longevity-genie/opengenes-mcp.git
cd opengenes-mcp
uv sync
Running the MCP Server
If you already cloned the repo you can run the server with uv:
# Start the MCP server locally (HTTP mode)
uv run server
# Or start in STDIO mode
uv run stdio
# Or start in SSE mode
uv run sse
Database Schema
Detailed schema information
Main Tables
- lifespan_change (47 columns): Experimental lifespan data with intervention details across model organisms
- gene_criteria (2 columns): Gene classifications by aging criteria (12 different categories)
- gene_hallmarks (2 columns): Hallmarks of aging mappings for genes
- longevity_associations (11 columns): Population genetics longevity data from human studies
Key Fields
- HGNC: Gene symbol (primary identifier across all tables)
- model_organism: Research organism (mouse, C. elegans, fly, etc.)
- effect_on_lifespan: Direction of lifespan change (increases/decreases/no change)
- intervention_method: Method of genetic intervention (knockout, overexpression, etc.)
- criteria: Aging-related gene classification (12 categories)
- hallmarks of aging: Biological aging processes associated with genes
Example Queries
Sample SQL queries for common research questions
-- Get top genes with most lifespan experiments
SELECT HGNC, COUNT(*) as experiment_count
FROM lifespan_change
WHERE HGNC IS NOT NULL
GROUP BY HGNC
ORDER BY experiment_count DESC
LIMIT 10;
-- Find genes that increase lifespan in mice
SELECT DISTINCT HGNC, effect_on_lifespan
FROM lifespan_change
WHERE model_organism = 'mouse'
AND effect_on_lifespan = 'increases lifespan'
AND HGNC IS NOT NULL;
-- Get hallmarks of aging for genes
SELECT HGNC, "hallmarks of aging"
FROM gene_hallmarks
WHERE "hallmarks of aging" LIKE '%mitochondrial%';
-- Find longevity associations by ethnicity
SELECT HGNC, "polymorphism type", "nucleotide substitution", ethnicity
FROM longevity_associations
WHERE ethnicity LIKE '%Italian%';
-- Find genes with both lifespan effects and longevity associations
SELECT DISTINCT lc.HGNC
FROM lifespan_change lc
INNER JOIN longevity_associations la ON lc.HGNC = la.HGNC
WHERE lc.HGNC IS NOT NULL;
Safety Features
- Read-only access: Only SELECT queries are allowed
- Input validation: Blocks INSERT, UPDATE, DELETE, DROP, CREATE, ALTER, TRUNCATE operations
- Error handling: Comprehensive error handling with informative messages
Testing & Verification
The MCP server is provided with comprehensive tests including LLM-as-a-judge tests that evaluate the quality of responses to complex queries. However, LLM-based tests are disabled by default in CI to save costs.
Environment Setup for LLM Agent Tests
If you want to run LLM agent tests that use MCP functions with Gemini models, you need to set up a .env file with your Gemini API key:
# Create a .env file in the project root
echo "GEMINI_API_KEY=your-gemini-api-key-here" > .env
Note: The .env file and Gemini API key are only required for running LLM agent tests. All other tests and basic MCP server functionality work without any API keys.
Running Tests
Run tests for the MCP server:
uv run pytest -vvv -s
You can also run manual tests:
uv run python test/manual_test_questions.py
You can use MCP inspector with locally built MCP server same way as with uvx.
Note: Using the MCP Inspector is optional. Most MCP clients (like Cursor, Windsurf, etc.) will automatically display the available tools from this server once configured. However, the Inspector can be useful for detailed testing and exploration.
If you choose to use the Inspector via npx, ensure you have Node.js and npm installed. Using nvm (Node Version Manager) is recommended for managing Node.js versions.
Example questions that MCP helps to answer
Research questions you can explore with this MCP server
- Interventions on which genes extended mice lifespan most of all?
- Which knockdowns were most lifespan extending on model animals?
- What processes are improved in GHR knockout mice?
- Which genetic intervention led to the greatest increase in lifespan in flies?
- To what extent did the lifespan increase in mice overexpressing VEGFA?
- Are there any liver-specific interventions that increase lifespan in mice?
- Which gene-longevity association is confirmed by the greatest number of studies?
- What polymorphisms in FOXO3 are associated with human longevity?
- In which ethnic groups was the association of the APOE gene with longevity shown?
- Is the INS gene polymorphism associated with longevity?
- What genes are associated with transcriptional alterations?
- Which hallmarks are associated with the KL gene?
- How many genes are associated with longevity in humans?
- What types of studies have been conducted on the IGF1R gene?
- What evidence of the link between PTEN and aging do you know?
- What genes are associated with both longevity and altered expression in aged humans?
- Is the expression of the ACE2 gene altered with aging in humans?
- What genes need to be downregulated in worms to extend their lifespan?
Contributing
We welcome contributions from the community! 🎉 Whether you're a researcher, developer, or enthusiast interested in aging and longevity research, there are many ways to get involved:
We especially encourage you to try our MCP server and share your feedback with us! Your experience using the server, any issues you encounter, and suggestions for improvement are incredibly valuable for making this tool better for the entire research community.
Ways to Contribute
- 🐛 Bug Reports: Found an issue? Please open a GitHub issue with detailed information
- 💡 Feature Requests: Have ideas for new functionality? We'd love to hear them!
- 📝 Documentation: Help improve our documentation, examples, or tutorials
- 🧪 Testing: Add test cases, especially for edge cases or new query patterns
- 🔍 Data Quality: Help identify and report data inconsistencies or suggest improvements
- 🚀 Performance: Optimize queries, improve caching, or enhance server performance
- 🌐 Integration: Create examples for new MCP clients or AI systems
- 🎥 Tutorials & Videos: Create tutorials, video guides, or educational content showing how to use MCP servers
- 📖 User Stories: Share your research workflows and success stories using our MCP servers
- 🤝 Community Outreach: Help us evangelize MCP adoption in the bioinformatics community
Tutorials, videos, and user stories are especially valuable to us! We're working to push the bioinformatics community toward AI adoption, and real-world examples of how researchers use our MCP servers (this one and others we develop) help demonstrate the practical benefits and encourage wider adoption.
Getting Started
- Fork the repository
- Create a feature branch (
git checkout -b feature/amazing-feature) - Make your changes and add tests
- Run the test suite (
uv run pytest) - Commit your changes (
git commit -m 'Add amazing feature') - Push to your branch (
git push origin feature/amazing-feature) - Open a Pull Request
Development Guidelines
- Follow the existing code style (we use
blackfor formatting) - Add tests for new functionality
- Update documentation as needed
- Keep commits focused and write clear commit messages
Questions or Ideas?
Don't hesitate to open an issue for discussion! We're friendly and always happy to help newcomers get started. Your contributions help advance open science and longevity research for everyone. 🧬✨
Known Issues
Database Coverage
Currently, this MCP server uses only a subset of the complete OpenGenes database. The full OpenGenes database contains additional tables and data that are not yet included in our MCP implementation. We need contributors to help extend support for the complete database! If you're interested in helping expand the database coverage, please see our Contributing section.
Test Coverage
While we provide comprehensive tests including LLM-as-a-judge evaluations, not all test cases have been manually verified against the actual OpenGenes web interface. Some automated test results may need manual validation to ensure accuracy. Contributions to improve test coverage and validation are welcome.
License
This project is licensed under the MIT License.
Acknowledgments
- OpenGenes Database for the comprehensive aging research data
- Rafikova E, Nemirovich-Danchenko N, Ogmen A, Parfenenkova A, Velikanova A, Tikhonov S, Peshkin L, Rafikov K, Spiridonova O, Belova Y, Glinin T, Egorova A, Batin M. Open Genes-a new comprehensive database of human genes associated with aging and longevity. Nucleic Acids Res. 2024 Jan 5;52(D1):D950-D962. doi: 10.1093/nar/gkad712. PMID: 37665017; PMCID: PMC10768108.
- Model Context Protocol for the protocol specification
- FastMCP for the MCP server framework
This project is part of the Longevity Genie organization, which develops open-source AI assistants and libraries for health, genetics, and longevity research.
Other MCP Servers by Longevity Genie
We also develop other specialized MCP servers for biomedical research:
- biothings-mcp - MCP server for BioThings.io APIs, providing access to gene annotation (mygene.info), variant annotation (myvariant.info), and chemical compound data (mychem.info)
We are supported by:
HEALES - Healthy Life Extension Society
and
IBIMA - Institute for Biostatistics and Informatics in Medicine and Ageing Research




